Spatial Moran process
The spatial Moran process models stochastic evolution in structured populations. Population structure is represented by graphs where each vertex represents an individual and edges define the neighbourhood of each individual. The graph can have any structure – for example, square lattices describe spatially extended systems, small-world networks (Watts & Strogatz, 1998) or scale-free networks (Barabasí & Albert, 1999) model social structures – and links between nodes may have different strengths. Links can even be directed such that one individual is a neighbour of another but not vice versa. A fully connected graph, where each node is equally linked to every other node, is essentially equivalent to an unstructured population. Mathematically, the structure of the graph is determined by the adjacency matrix \(\mathbf{W} = [w_{ij}]\) where \(w_{ij}\) denotes the strength of the link between nodes \(i\) and \(j\). If \(w_{ij} =0\) and \(w_{ji} =0\) then the two nodes \(i\) and \(j\) are not connected.
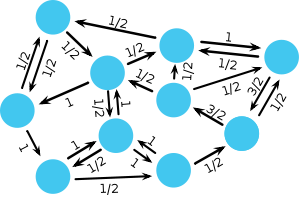
The essential difference to the original Moran process that arises through the population structure is that the offspring of the focal individual replaces a random neighbor instead of a random member of the population. An additional minor difference is that the offspring cannot replace the focal individual but, in principle, this can be implemented by adding loops to each node.
Provided that the graph is connected, i.e. each node is connected to every other node through a series of links, the population maintains the same two absorbing states with all residents or all mutants. If the graph is not connected, mutant fixation is impossible. As before, we are interested in the fixation probability of a single mutant in a structured resident population and, more specifically, the effect of limited dispersal of offspring on the mutant fixation probability.
Evolutionary dynamics
In the simplest case, the population is composed of residents (blue) and mutants (orange):
Setup: The state of the population is given by the configuration of residents (blue, fitness \(1\)) and mutants (orange, fitness \(r\)) on a graph. In this sample graph all links are undirected (bi-directional).
Step 1: At each time step a focal individual is chosen for reproduction with a probability proportional to its fitness (here, a mutant is selected for reproduction).
Step 2: A randomly chosen 'neighbour' of the focal individual (vivid colours) is eliminated (here, a resident is selected for death).
Step 3: The offspring replaces the eliminated individual.
As in the original Moran process, the above steps are repeated until either one of the two absorbing states is reached: a homogeneous state of either all residents (mutant went extinct) or all mutants (resident displaced). No other stable equilibrium state is possible provided that the graph is connected
Fixation probability
In contrast to unstructured (well-mixed) populations, the state of the population is not only determined by just the number of mutants (or residents) but rather needs to take the spatial distribution of mutants and residents into account. Rather surprisingly, however, it turns out that for a large class of graphs, called circulations, the spatial distribution of mutants and residents does not affect their fixation probabilities. This forms the basis for the conjecture by Maruyama (1970) and Slatkin (1981) speculating that population structure leaves fixation probabilities unchanged. Indeed this is true for structures as diverse as complete graphs (every vertex is connected to every other vertex), lattices or random regular graphs (see e.g. Bollobás, 1995). Further examples and interactive demonstrations are covered in Moran graphs.
Circulation graphs
In order to characterize the class of graphs that lead to the same fixation probability \(\rho_1\) as the original Moran process in unstructured populations, let us introduce the flux through each node. The sum of the weights of the incoming links \(f_i^\text{in}=\sum_j w_{ji}\) denotes the flux entering node \(i\). \(f_i^\text{in}\) relates to a temperature because it indicates the rate at which the occupant of node \(i\) gets replaced. ‘Hot’ nodes are frequently replaced and ‘cold’ nodes only rarely change their type. In analogy, the sum of the weights of the outgoing links \(f_i^\text{out}=\sum_j w_{ij}\) denotes the flux leaving node \(i\). \(f_i^\text{out}\) determines the impact of node \(i\) on its neighborhood. The matrix \(\mathbf{W}=[w_{ij}]\) is a circulation if \(f_i^\text{in}=f_i^\text{out}\) holds for all nodes.
- Circulation Theorem: The Moran process on a graph results in the same fixation probability \(\rho_1\) of a single mutant as in an unstructured population if the graph is a circulation.
At any point in time during the invasion process of mutants on a circulation graph, it is possible to identify connected subsets of nodes on the graph that are occupied by mutants such that all adjacent nodes of each subset are occupied by residents. Obviously, the state of the population changes only if a replacement occurs along one of the links connecting residents and mutants (see figure). Multiple such subsets may exist and, in fact, the evolutionary process may split large connected subsets of mutants into two smaller ones or may merge two previously unconnected subsets into one larger subset. For each subset, the circulation theorem establishes that the sum of the weights of links pointing out of the subset (connecting a mutant node to an adjacent resident node) equals the sum of the weights of links pointing into the subset (connecting an adjacent resident with a mutant node within the subset). Since the influx is balanced by the outflux, \(f_i^\text{in}=f_i^\text{out}\), for all nodes, increasing the mutant subset by replacing an adjacent resident node with a mutant or decreasing the subset by replacing a mutant with a resident, does not affect the flux balance of the subset. For the same reason, the balance remains unchanged if subsets merge or if one subset splits into two. Recall that the number of mutants in the population changes only if a replacement occurs along any link that connects residents with mutants or vice versa. Because the Moran process essentially selects links with a probability proportional to the link weight and the fitness of the node at its tail, it follows that for each subset, the probability that another mutant is added is simply given by \(r/(1 + r)\) and the complementary probability that one is removed is \(1/(1 + r)\). Since this holds for each subset, it also holds for the entire population and is independent of the number, size, shape and distribution of mutant subsets. This invariance applies if and only if the circulation theorem is satisfied. Consequentially, the fixation probability on circulation graphs reduces to the recursive equation derived for the original Moran process.
Note that even though the fixation probabilities remain unchanged on circulation graphs, the corresponding fixation times are very sensitive to the details of the population structure and pose a much harder problem. The circulation theorem only ensures that the ratio of the transition probabilities \(T^+/T^− = r\) remains unchanged but even on circulation graphs \(T^+\) and \(T^−\) depend not only on the number but also on the distribution of mutants. Generally, population structures tend to substantially increase the fixation times because the structure limits the possibilities for mutants to conquer new nodes.
Finally, the circulation theorem indirectly suggests that some population structures do affect the fixation probabilities. Thus it becomes an intriguing question whether population structures may act as evolutionary suppressors that suppress selection and enhance random drift (\(\rho<\rho_1\) for \(r > 1\)) or, conversely, as evolutionary amplifiers that enhance selection and suppresses random drift (\(\rho>\rho_1\) for \(r > 1\))?
Publications
- Lieberman, E., Hauert, C. & Nowak, M. (2005) Evolutionary dynamics on graphs Nature 433 312-316 doi: 10.1038/nature03204.
References
- Barabasí, A.-L. & Albert, R. (1999) Emergence of scaling in random networks Science 286 509–512.
- Bollobás, B. (1995) Random Graphs, New York, Academic.
- Maruyama, T. (1974) A Simple Proof that Certain Quantities are Independent of the Geographical Structure of Population Theor. Pop. Biol. 5 148-154.
- Slatkin, M. (1981) Fixation probabilities and fixation times in a subdivided population Evolution 35 477-488.
- Watts, D. J. & Strogatz, S. H. (1998) Collective dynamics of 'small world' networks Nature 393 440–442.